DavidStables (talk | contribs) No edit summary |
DavidStables (talk | contribs) m (DavidStables moved page Discovery health information model to Health information model) |
||
| (177 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
This article describes the approach taken to producing information models, including ; what they are, what their purpose is, and what the technical components of the models are. | |||
The article does not include the content of any particular model. | |||
== What is the health information model (IM) and what is its purpose? == | |||
The IM is a representation of the meaning and structure of data held in the electronic records of the health and social care sector, together with libraries of query, value sets, concept sets, data set definitions and mappings. | |||
The main purpose is to bridge the chasm that exists between highly technical digital representations and plain language so that when questions are asked of data, a lay person could use plain language without prior knowledge of the underlying models. | |||
It is a computable abstract logical model, not a physical structure or schema. "computable" means that operational software operates directly from the model artefacts, as opposed to using the model for illustration purposes. As a logical model it models data that may be physically held any a variety of different types of data stores, including relational or graph data stores. Because the model is independent of the physical schemas, the model itself has to be interoperable and without any proprietary lock in. | |||
The IM is a broad model that integrates a set of different approaches to modelling using a common ontology. The components of the model are: | |||
The | # A set of ontologies, which is a vocabulary and definitions of the concepts used in healthcare, or more simply put, a vocabulary of health. The ontologies is made up of the world's leading ontology Snomed-CT, with a London extensions, various code based taxonomies (e.g. ICD10, Read, supplier codes and local codes) | ||
# A common data model, which is a set of classes and properties, using the vocabulary, that represent the data and relationships as published by live systems that have published data, Note that this data model is NOT a standard model but a collated set of entities and relationships bound to the concepts based on real data, that are mapped to a common model. | |||
# A library of business specific concept value sets, (aka reference sets) which are expression constraints on the ontology for the purpose of query | |||
# A catalogue of reference data such as geographical areas, organisations and people derived and updated from public resources. | |||
# A library of Data set (query) definitions for querying and extracting instance data from the information model, reference data, or health records. | |||
# A set of maps creating mappings between published concepts and the core ontology as well as structural mappings between submitted data and the data model. | |||
# An open source set of utilities that can be used to browse, search, or maintain the model. | |||
<br /> | |||
== | == Model building blocks and visualisation == | ||
The | The model consists of classes, sets and objects that are instances of classes. | ||
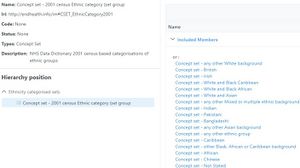
[[File:Ethnicity.jpg|thumb|Ethnicity]] | |||
Objects can act as objects in their own rights (e.g. an instance of chest pain) or may also act as classes (e.g. the class of objects that are chest pain). Likewise sets have members that are objects and the objects may also act as classes or sets. For example a set for the 2011 Ethnicity census will contain a member object of "British" which is also a set with members such as English and so on. | |||
The model itself is stored as an RDF based knowledge graph, which means it is implementable in any mainstream Graph database technology. There are no vendor specific extensions to RDF. | |||
In line with the RDF standard, all persistent types, classes, , property identifiers and object value identifiers are uniquely named using international resource identifiers. In most cases the identifiers are externally provided (e.g. Snomed-CT identifiers) whilst in others that have been created for a particular model. Organisations that author elements of the models use their own identifiers. | |||
From a data modelling perspective the arrangements of types may be referred to as archetypes, which are conceptually similar to FHIR profiles. In the semantic web world they would be considered "shapes". There are an unlimited number of these which frees the model from any particular conventional relational database schema. Inheritance of types is supported which enables broad classifications of types and re-usability. | |||
The variation between the parts of the model that model terminology concepts and those that model data use slightly different grammars in keeping with their different purposes. The information model language describes the differences. | |||
The models can be viewed in their raw technical form (in JSON or Turtle) or can be viewed by the information model viewer at the online tool [https://im.endeavourhealth.net/#/ Information model directory] | |||
== Information model language == | |||
''Main article'' [[Health Information modelling language - overview|information modelling language]] describes the language in more detail. | |||
The semantic web approach is adopted for the purposes of identifiers and grammar. In this approach, data can be described via the use of a plain language grammar consisting of a subject, a predicate, and an object; A triple, with an additional context referred to as a graph or RDF data set. The theory is that all health data can be described in this way (with predicates being extended to include functions). | |||
However, the semantic web languages are highly complex and a set of more pragmatic approaches are taken for the more specialised structures. | |||
The consequence of this approach is that W3C web standards can be used such as the use of [[wikipedia:Resource_Description_Framework|Resource Descriptor Framework o]]<nowiki/>r RDF. This sees the world as a set of triples (subject/ predicate/ object) with some things named and somethings anonymous. Systems that adopt this approach can exchange data in a way that the semantics can be preserved. Whilst RDF is an incredibly arcane language at a machine level, the things it can describe can be very intuitive when represented visually. In other words the Information modelling approach involves an RDF Graph. | |||
In addition to semantic web languages, other commonly used languages are in place are used to enable the model to be accessed by more people.. For example the Snomed-CT expression constraint language is a common way of defining concept sets. ECL is logically equivalent to a closed world query on an open world OWL ontology. The IM language uses the semantic language of SPARQL together with entailment to model ECL but ECL can be exported or used as input as an alternative. | |||
<br /> | <br /> | ||
Latest revision as of 10:28, 21 August 2022
This article describes the approach taken to producing information models, including ; what they are, what their purpose is, and what the technical components of the models are.
The article does not include the content of any particular model.
What is the health information model (IM) and what is its purpose?
The IM is a representation of the meaning and structure of data held in the electronic records of the health and social care sector, together with libraries of query, value sets, concept sets, data set definitions and mappings.
The main purpose is to bridge the chasm that exists between highly technical digital representations and plain language so that when questions are asked of data, a lay person could use plain language without prior knowledge of the underlying models.
It is a computable abstract logical model, not a physical structure or schema. "computable" means that operational software operates directly from the model artefacts, as opposed to using the model for illustration purposes. As a logical model it models data that may be physically held any a variety of different types of data stores, including relational or graph data stores. Because the model is independent of the physical schemas, the model itself has to be interoperable and without any proprietary lock in.
The IM is a broad model that integrates a set of different approaches to modelling using a common ontology. The components of the model are:
- A set of ontologies, which is a vocabulary and definitions of the concepts used in healthcare, or more simply put, a vocabulary of health. The ontologies is made up of the world's leading ontology Snomed-CT, with a London extensions, various code based taxonomies (e.g. ICD10, Read, supplier codes and local codes)
- A common data model, which is a set of classes and properties, using the vocabulary, that represent the data and relationships as published by live systems that have published data, Note that this data model is NOT a standard model but a collated set of entities and relationships bound to the concepts based on real data, that are mapped to a common model.
- A library of business specific concept value sets, (aka reference sets) which are expression constraints on the ontology for the purpose of query
- A catalogue of reference data such as geographical areas, organisations and people derived and updated from public resources.
- A library of Data set (query) definitions for querying and extracting instance data from the information model, reference data, or health records.
- A set of maps creating mappings between published concepts and the core ontology as well as structural mappings between submitted data and the data model.
- An open source set of utilities that can be used to browse, search, or maintain the model.
Model building blocks and visualisation
The model consists of classes, sets and objects that are instances of classes.
Objects can act as objects in their own rights (e.g. an instance of chest pain) or may also act as classes (e.g. the class of objects that are chest pain). Likewise sets have members that are objects and the objects may also act as classes or sets. For example a set for the 2011 Ethnicity census will contain a member object of "British" which is also a set with members such as English and so on.
The model itself is stored as an RDF based knowledge graph, which means it is implementable in any mainstream Graph database technology. There are no vendor specific extensions to RDF.
In line with the RDF standard, all persistent types, classes, , property identifiers and object value identifiers are uniquely named using international resource identifiers. In most cases the identifiers are externally provided (e.g. Snomed-CT identifiers) whilst in others that have been created for a particular model. Organisations that author elements of the models use their own identifiers.
From a data modelling perspective the arrangements of types may be referred to as archetypes, which are conceptually similar to FHIR profiles. In the semantic web world they would be considered "shapes". There are an unlimited number of these which frees the model from any particular conventional relational database schema. Inheritance of types is supported which enables broad classifications of types and re-usability.
The variation between the parts of the model that model terminology concepts and those that model data use slightly different grammars in keeping with their different purposes. The information model language describes the differences.
The models can be viewed in their raw technical form (in JSON or Turtle) or can be viewed by the information model viewer at the online tool Information model directory
Information model language
Main article information modelling language describes the language in more detail.
The semantic web approach is adopted for the purposes of identifiers and grammar. In this approach, data can be described via the use of a plain language grammar consisting of a subject, a predicate, and an object; A triple, with an additional context referred to as a graph or RDF data set. The theory is that all health data can be described in this way (with predicates being extended to include functions).
However, the semantic web languages are highly complex and a set of more pragmatic approaches are taken for the more specialised structures.
The consequence of this approach is that W3C web standards can be used such as the use of Resource Descriptor Framework or RDF. This sees the world as a set of triples (subject/ predicate/ object) with some things named and somethings anonymous. Systems that adopt this approach can exchange data in a way that the semantics can be preserved. Whilst RDF is an incredibly arcane language at a machine level, the things it can describe can be very intuitive when represented visually. In other words the Information modelling approach involves an RDF Graph.
In addition to semantic web languages, other commonly used languages are in place are used to enable the model to be accessed by more people.. For example the Snomed-CT expression constraint language is a common way of defining concept sets. ECL is logically equivalent to a closed world query on an open world OWL ontology. The IM language uses the semantic language of SPARQL together with entailment to model ECL but ECL can be exported or used as input as an alternative.