Health Information modelling language - overview: Difference between revisions
DavidStables (talk | contribs) No edit summary |
DavidStables (talk | contribs) No edit summary |
||
| Line 6: | Line 6: | ||
Each sublanguage is either a single recognised standards based language, or is an amalgam of other known languages or structural concepts, these having been selected as the ones that are the closest fit to the information requirements that the Discovery health information model is designed to support. A single grammar and optional single syntax enables the model to operate in an integrated manner, but at the same time enables the sublanguages to be represented in their native standard languages. | Each sublanguage is either a single recognised standards based language, or is an amalgam of other known languages or structural concepts, these having been selected as the ones that are the closest fit to the information requirements that the Discovery health information model is designed to support. A single grammar and optional single syntax enables the model to operate in an integrated manner, but at the same time enables the sublanguages to be represented in their native standard languages. | ||
Like [https://www.nlm.nih.gov/research/umls/index.html UMLS], the sublanguages use a common link in cross reference, a [[ | Like [https://www.nlm.nih.gov/research/umls/index.html UMLS], the sublanguages use a common link in cross reference, a [[Discovery semantic ontology language|concept,]] which is identified with a unique identifier (Internationalised resource identifier - IRI) . a concept is usually named and defined semantically, and forms the means of traversing the model from different starting points to different end points, for different purposes. | ||
== Why another language? == | == Why another language? == | ||
| Line 31: | Line 31: | ||
The grammar for the semantic ontology language used for the Discovery ontology is [https://www.w3.org/TR/owl2-profiles/#OWL_2_EL OWL EL]. | The grammar for the semantic ontology language used for the Discovery ontology is [https://www.w3.org/TR/owl2-profiles/#OWL_2_EL OWL EL]. | ||
As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full | As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full information modelling language. | ||
== Value set language == | == Value set language == | ||
''Main article :'' [[Value sets]] | ''Main article :'' [[Value sets]] | ||
The language | The language uses Discovery syntax to model the value set itself with its member expressions supporting Discovery syntax, [https://www.w3.org/TR/owl2-syntax/ OWL2 functional syntax], and [https://confluence.ihtsdotools.org/display/DOCECL/Expression+Constraint+Language+-+Specification+and+Guide Expression constraint language] | ||
== Data model language == | == Data model language == | ||
Revision as of 09:30, 26 May 2020
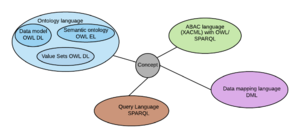
The Discovery Information Modelling Language (or DIMOL for short) is a mixed language that subsumes 5 sublanguages, brought together under a common grammar and an optional syntax.
The purpose of the language is to define a health information information model in a way that supports implementations of the model using different data base technologies and query languages.
Each sublanguage is either a single recognised standards based language, or is an amalgam of other known languages or structural concepts, these having been selected as the ones that are the closest fit to the information requirements that the Discovery health information model is designed to support. A single grammar and optional single syntax enables the model to operate in an integrated manner, but at the same time enables the sublanguages to be represented in their native standard languages.
Like UMLS, the sublanguages use a common link in cross reference, a concept, which is identified with a unique identifier (Internationalised resource identifier - IRI) . a concept is usually named and defined semantically, and forms the means of traversing the model from different starting points to different end points, for different purposes.
Why another language?
There are hundreds of computer languages and thousands of natural languages, many of which are accepted as "standards" in many communities. Why have another?
Within the health informatics community, a historical separation has evolved between two modelling camps, those that model semantics of concepts via an ontology (aka terminologists), and those that model data structures for storing and transmitting data (aka structuralists). This separation reflects the difference in purpose, the difference in mindset, and the difference in skills required by the different disciplines.
A problem with this separation occurs at the points of overlap between the two: e.g. properties and values. Both camps model their classes with properties and ranges (the allowable values of properties). In many cases, properties can be modelled either within the semantic space, or within the structural space, or both.
For example, it is possible to model a surgical operation as a data structure with a body site attribute. (FHIR R4 procedure) does precisely this. It is equally possible to model a procedure by including the body site either as a qualifier of a type of procedure, or as part of the procedure definition itself. Snomed-CT does precisely this. Both approaches can use the same concept for the body site itself, but they would use separate property concepts for the property of "has body site" itself. This separation of approach can lead to massive divergences. Taking the structuralist approach and extending it results in archetypes of the kind modelled by OpenEHR. Taking the ontological approach further leads to complex nested expressions which are nigh on impenetrable.
Having a grammar and syntax that encompasses both semantics and structure, makes the use of the common overlapping concepts much easier to manage. A classic structural concept such as an encounter record and its semantic definition, can be seamlessly integrated. The use of a current language is one option.
For example, it is possible to model data in OWL2 DL by extensive use of complex OWL constructs including functional properties, property domains, ranges, precise cardinality. It is also possible to model query as OWL expressions, except for function parameters. However, the purpose of OWL is to support reasoning, and reasoners use the open world assumption.. Data models and data query uses a closed world assumption and query languages are declarative in nature i.e. instructions as to what to do. Using OWL for purposes other than reasoning is like using English to prove Pythagoras theorem.
Thus is seems sensible to keep these separate. However, in doing so the separation continues and as time goes on there is some divergence. Bringing them together, at least as a temporary measure to solve a particular set of information requirements, seems worthwhile. Hence a new language.
Semantic ontology language
Main article Discovery semantic ontology language
The semantic ontology language is part of the Discovery data modelling language.
The grammar for the semantic ontology language used for the Discovery ontology is OWL EL.
As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full information modelling language.
Value set language
Main article : Value sets
The language uses Discovery syntax to model the value set itself with its member expressions supporting Discovery syntax, OWL2 functional syntax, and Expression constraint language
Data model language
This part of the language is used to define a data model. Data model content may be automatically generated from the OWL2 representation of the data model, with the result that a much simpler syntax can be used. The data modelling language relies on the OWL2 language for managing subclasses, sub properties, domains, ranges, cardinality. Thus in Discovery the language is really just a simplified output syntax from the more sophisticated description logic.
Ontology purists will notice that modelling a data model in OWL2 is in fact a breach of the fundamental open world assumption view of the world taken in ontologies and instead applies the https://en.wikipedia.org/wiki/Closed-world_assumption view instead. Conseqently, a data model would normally be used independently of DL reasoners, and therefore a syntax that seperates the two is provided.
Data definition language (query)
This language is designed to define the building of data sets from an underlying data model and semantic ontology, supplemented with value sets defined using the value set class, itself modelled in OWL2 syntax.
Schema implementation mapping
This part of the language is used to define mappings between the data model and an actual schema to enable query and filers to automatically cope with the ever extending ontolofy and data properties.
The language can be used to auto generate starter schemas for implementation i.e. schemas that will then be optimised for real world use.
One significant mapping capability is the use of the entity subtype attribute which is the means by which a relational model with triple extensions can cope with any extension to a core data model entity via subtyping or subcomponent constraining.