Health Information modelling language - overview: Difference between revisions
DavidStables (talk | contribs) No edit summary |
DavidStables (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
The Discovery | The Discovery Data Modelling Language (or ''DIMOL'' for short) is a [[wikipedia:Mixed_language|mixed language]] that subsumes 4 [[wikipedia:Sublanguage|sublanguages]], brought together under a common grammar and set of optional syntaxes. | ||
The language is a | The purpose of the language is to define a health information information model in a way that supports implementations of the model using different data base technologies and query languages. | ||
Each sublanguage is either a single recognised standards based language, or is an amalgam of other known languages or structural concepts, these having been selected as the ones that are the closest fit to the information requirements that an information model is designed to support. A single grammar and optional single syntax enables the model to operate seamlessly and in a fully integrated manner. | |||
The sublanguages share a common 'link' item, the concept. | |||
== Background to the language and standards == | == Background to the language and standards == | ||
Revision as of 08:18, 25 May 2020
The Discovery Data Modelling Language (or DIMOL for short) is a mixed language that subsumes 4 sublanguages, brought together under a common grammar and set of optional syntaxes.
The purpose of the language is to define a health information information model in a way that supports implementations of the model using different data base technologies and query languages.
Each sublanguage is either a single recognised standards based language, or is an amalgam of other known languages or structural concepts, these having been selected as the ones that are the closest fit to the information requirements that an information model is designed to support. A single grammar and optional single syntax enables the model to operate seamlessly and in a fully integrated manner.
The sublanguages share a common 'link' item, the concept.
Background to the language and standards
The modelling language draws heavily on 3 main standards; The W3C OWL2 DL, W3C SPARQL languages and UML.
Language ideas have been supplemented by CYPHER (property graph) and SQL, as that is likely to be the run time query language commonly used.
The approach to syntax has been to offer a standard language where users prefer, but also to have a simple JSON based syntax (Discovery syntax) that many will find easier to follow. There are converters to standard languages available where relevant.
Semantic ontology
The semantic ontology language is part of the Discovery information modelling language.
The language used for the Discovery ontology is an OWL2 DL profile with some limitations making it OWL2 RL with the addition of exact cardinality (for use in the closed world data model), and Object union in ranges for use in value sets.
An ontology is concerned with the meaning of things and defines the meaning of things in a way that allows inferences to be made about other things. An ontology designed with a language such as OWL2 allows machines to make the sophisticated inferences required to support query and decision support. OWL2 itself is based on an underlying Description Logic variation which have underpinned machine reasoning for decades.
Ontologies have been used in health information systems for many years and more recently, the emergence of Snomed-CT as the de-facto health terminology has illustrated the potential power of description logic which underpins OWL.
Discovery implementation supports the official OWL2 functional syntax but also provides a JSON based syntax, “Discovery Syntax” which absorbs other language constructs for other parts of the model, and thus can also be used for the language of all of the IM components.
The Ontology language describes four main structural types which cut across the information model content parts as described above, which are:
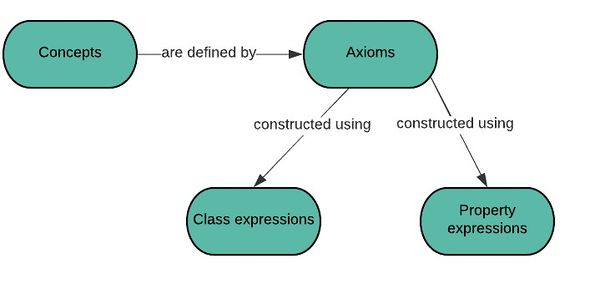
Concepts classes and properties
main article : Concepts classes and properties form the building blocks used by the language and are declared in an ontology.
Value sets
Main article : Value sets
The language supports three forms for representing value sets; OWL2 Discovery syntax, OWL2 functional syntax, and Expression constraint language
Data model
This part of the language is used to define a data model. Data model content may be automatically generated from the OWL2 representation of the data model, with the result that a much simpler syntax can be used. The data modelling language relies on the OWL2 language for managing subclasses, sub properties, domains, ranges, cardinality. Thus in Discovery the language is really just a simplified output syntax from the more sophisticated description logic.
Ontology purists will notice that modelling a data model in OWL2 is in fact a breach of the fundamental open world assumption view of the world taken in ontologies and instead applies the https://en.wikipedia.org/wiki/Closed-world_assumption view instead. Conseqently, a data model would normally be used independently of DL reasoners, and therefore a syntax that seperates the two is provided.
Data set definition language
This language is designed to define the building of data sets from an underlying data model and semantic ontology, supplemented with value sets defined using the value set class, itself modelled in OWL2 syntax.
Schema implementation mapping
This part of the language is used to define mappings between the data model and an actual schema to enable query and filers to automatically cope with the ever extending ontolofy and data properties.
The language can be used to auto generate starter schemas for implementation i.e. schemas that will then be optimised for real world use.
One significant mapping capability is the use of the entity subtype attribute which is the means by which a relational model with triple extensions can cope with any extension to a core data model entity via subtyping or subcomponent constraining.