Health Information modelling language - overview: Difference between revisions
DavidStables (talk | contribs) No edit summary |
DavidStables (talk | contribs) |
||
| Line 6: | Line 6: | ||
It is not necessary to understand the languages used in order to understand the modelling, but for those who have an interest, and have a technical aptitude, and do not want to need to grips with the fundamentals of logic the best places to start are with [https://www.w3.org/TR/owl2-primer/ OWL2], [https://graphql.org/ GRAPHQL] and [[wikipedia:XACML|ABAC.]] For those who want to get to grips with underlying logic, the best place to start is First order Logic, Description logic, High order logic and an understanding of at least one programming language like C# Java, Java script, Python etc + any query language such as SQL. | It is not necessary to understand the languages used in order to understand the modelling, but for those who have an interest, and have a technical aptitude, and do not want to need to grips with the fundamentals of logic the best places to start are with [https://www.w3.org/TR/owl2-primer/ OWL2], [https://graphql.org/ GRAPHQL] and [[wikipedia:XACML|ABAC.]] For those who want to get to grips with underlying logic, the best place to start is First order Logic, Description logic, High order logic and an understanding of at least one programming language like C# Java, Java script, Python etc + any query language such as SQL. | ||
The | == The roles of each language == | ||
=== OWL2 === | |||
OWL2, like Snomed-CT, forms the logical basis for the static data representations, including semantic definition, data modelling and modelling of value sets. | |||
Because raw OWL2 language (with its 4 syntaxes) is quite arcane to use, Discovery has created a JSON/XML based projection of the language which is simpler to follow and use. This makes access to the information model building blocks more interoperable, for example via the use of REST APIs and JSON. The JSON is mapped directly to classes to enable processing via languages such as Java. RDF/XML (one of the OWL syntaxes) can be exchanged as an option but many are unfamiliar with the OWL variation. | |||
In its usual use, OWL2 is used for reasoning and classification via the use of the [[wikipedia:Open-world_assumption|Open world assumption]]. This is also used in the information modelling, but the need to constrain the chaos of health data via data models requires the use of a [[wikipedia:Closed-world_assumption|closed world assumption.]] To enable this the OWL syntax is still used but is interpreted in a closed world manner. For example, where OWL2 models domains of a property in order to infer the class of a certain entity, Discovery uses the same syntax for use in editorial policies. Where OWL2 may say that one of the domains of a causative agent is an allergy (i.e.an unknown class with a property of causative agent is likely to be an allergy), in the data modelling the editorial policy states that an allergy can only have properties that are allowed via the property domain. Thus the Snomed MRCM can be modelled in OWL2. | |||
=== GRAPHQL === | |||
Graph QL , despite its name is not in itself a query language but a way of representing the graph like structure of a underlying model that has been built using OWL. GRAPH QL has a very simple class property representation, is ideal for REST APIs and results are JSON objects in line with the approach taken by the above Discovery syntax. | |||
Nevertheless, GRAPHQL considers properties to be functions (high order logic) and therefore properties can accept parameters. For example, a patient's average systolic blood pressure reading could be considered a property with a single parameter being a list of the last 3 blood pressure readings. | |||
Thus GRAPHQL capability is extended by enabling property parameters as types to support such things as filtering, sorting and limiting in the same way as any other query language by modelling types passed as parameters. Subqueries are then supported in the same way. | |||
GRAPH QL has been chosen over SPARQL for reasons of simplicity and may now consider GRAPHQL to be a de-facto standard. | |||
== ABAC language == | |||
''Main article :'' [[Discovery ABAC language|Discovery ABAC language ]] | |||
The standard [[wikipedia:XACML|XACML]] specifies a language that may be used to implement ABAC. XACML includes a set of grammatical concepts such as policy sets, policies, rules, combination rules, targets, obligations, effects and so on with many and variable sophisticated tokens and functions used to build the policy rules. XACML has its own XML syntax that can be used directly. | |||
This language is somewhat disconnected with the other standards in terms of syntax and approach to vocab. Consequently Discovery uses a J[[Discovery ABAC language|SON profile of XACML]] as its ABAC language which itself models the attributes as OWL properties, and uses SPARQL as its rule representation. | |||
== Objectives and purposes == | |||
The purpose of the language is to help build a health information information model in a way that supports implementations of the model using different data base technologies and query languages. | The purpose of the language is to help build a health information information model in a way that supports implementations of the model using different data base technologies and query languages. | ||
The underlying philosophy behind the language is described in the article : [[Information modelling language - philosophy]] | The underlying philosophy behind the use language is described in the article : [[Information modelling language - philosophy]] | ||
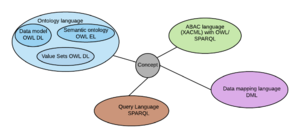
[[File:Sublanguages.png|thumb]] | [[File:Sublanguages.png|thumb]] | ||
This article focuses on the description of the language itself. | This article focuses on the description of the language itself. | ||
As mentioned above each sublanguage is based on the grammar of a single recognised standards based language, the language having been selected as the ones that are the closest fit to the information requirements that the Discovery health information model is designed to support. A single grammar and optional single syntax enables the model to operate in an integrated manner, but at the same time enables the sublanguages to be represented in their native standard languages. | |||
Like [https://www.nlm.nih.gov/research/umls/index.html UMLS], the sublanguages use a common link in cross reference, a [[Discovery semantic ontology language|concept,]] which is identified with a unique identifier (Internationalised resource identifier - IRI) . a concept is usually named and defined semantically, and forms the means of traversing the model from different starting points to different end points, for different purposes. | Like [https://www.nlm.nih.gov/research/umls/index.html UMLS], the sublanguages use a common link in cross reference, a [[Discovery semantic ontology language|concept,]] which is identified with a unique identifier (Internationalised resource identifier - IRI) . a concept is usually named and defined semantically, and forms the means of traversing the model from different starting points to different end points, for different purposes. | ||
== Ontology | == Semantic Ontology == | ||
''Main article'' [[Discovery semantic ontology language]] | ''Main article'' [[Discovery semantic ontology language]] | ||
| Line 28: | Line 52: | ||
As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full information modelling language. | As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full information modelling language. | ||
Together with the query language, OWL2 DL makes the language compatible also with [https://confluence.ihtsdotools.org/display/DOCECL/Expression+Constraint+Language+-+Specification+and+Guide Expression constraint language] which is used as the standard for specifying Snomed-CT expression query | Together with the query language, OWL2 DL makes the language compatible also with [https://confluence.ihtsdotools.org/display/DOCECL/Expression+Constraint+Language+-+Specification+and+Guide Expression constraint language] which is used as the standard for specifying Snomed-CT expression query. | ||
Ontology purists will notice that modelling a data model in OWL2 is in fact a breach of the fundamental [[wikipedia:Open-world_assumption|open world assumption]] view of the world taken in ontologies and instead applies the | Ontology purists will notice that modelling a data model in OWL2 is in fact a breach of the fundamental [[wikipedia:Open-world_assumption|open world assumption]] view of the world taken in ontologies and instead applies the [[wikipedia:Closed-world_assumption|closed world assumption]] view instead. Consequently, a data model would normally be used independently of DL | ||
The ontologies that are modelled are considered as modular ontologies. it is not expected that one "mega ontology" would be authored but that there would be maximum sharing of concept definitions (known as axioms) which results in a super ontology of modular ontologies. | |||
== Data | == Data modelling and semantic interoperability == | ||
Data models, and concept definitions and objects are modelled using the Graph paradigm. As a result, all content can be viewed as [[wikipedia:Semantic_triple|semantic triples]] consisting of subject predicate and object. | Data models, and concept definitions and objects are modelled in OQL language using the Graph paradigm. As a result, all content can be viewed as [[wikipedia:Semantic_triple|semantic triples]] consisting of subject predicate and object. | ||
A | Data Modelling takes account of ontology modularisation. A particular data model is a particular business oriented perspective on a set of concepts. As there are potentially thousands of different perspectives (e.g. a GP versus a geneticist) there are potentially unlimited number of data models. All the data models in Discovery share the same atomic concepts and same semantic definition across ontologies where possible, but where not, mapping relationships are used. The binding of a data model to its property values is based on a business specific model. For example a standard FHIR resource will map directly to the equivalent data model class, property and value set, whose meaning is defined in the semantic ontology, but the same data may be carried in a non FHIR resource without loss of interoperability. | ||
A common approach to modelling and use of a standard approach to ontology, together with modularisation, means that any sending or receiving machine which uses concepts from the "super" ontology can adopt full semantic interoperability. If both machines use the same data model for the same business, the data may presented in the same relationship, but if the two machines use different data models for different businesses they may present the data in different ways but without any loss of meaning or query capability | |||
== Data mapping sublanguage == | == Data mapping sublanguage == | ||
| Line 50: | Line 74: | ||
the main use case for he mapping sublanguage is data transformation. This uses techniques such as [[wikipedia:Object-relational_mapping|Object relational mapping]] and therefore the transform instructions in the form of maps, follow this approach. There is no single standard for ORM maps but best practice of the kind supported by open source utilities such as Hibernate is followed: | the main use case for he mapping sublanguage is data transformation. This uses techniques such as [[wikipedia:Object-relational_mapping|Object relational mapping]] and therefore the transform instructions in the form of maps, follow this approach. There is no single standard for ORM maps but best practice of the kind supported by open source utilities such as Hibernate is followed: | ||
<br /> | |||
Revision as of 10:56, 8 December 2020
Background and rationale
Yet another language?
No. The health information modelling languages described herein are in fact a small number of open standard languages, configured and represented to make the models easier to use and more useful in the real world.
It is not necessary to understand the languages used in order to understand the modelling, but for those who have an interest, and have a technical aptitude, and do not want to need to grips with the fundamentals of logic the best places to start are with OWL2, GRAPHQL and ABAC. For those who want to get to grips with underlying logic, the best place to start is First order Logic, Description logic, High order logic and an understanding of at least one programming language like C# Java, Java script, Python etc + any query language such as SQL.
The roles of each language
OWL2
OWL2, like Snomed-CT, forms the logical basis for the static data representations, including semantic definition, data modelling and modelling of value sets.
Because raw OWL2 language (with its 4 syntaxes) is quite arcane to use, Discovery has created a JSON/XML based projection of the language which is simpler to follow and use. This makes access to the information model building blocks more interoperable, for example via the use of REST APIs and JSON. The JSON is mapped directly to classes to enable processing via languages such as Java. RDF/XML (one of the OWL syntaxes) can be exchanged as an option but many are unfamiliar with the OWL variation.
In its usual use, OWL2 is used for reasoning and classification via the use of the Open world assumption. This is also used in the information modelling, but the need to constrain the chaos of health data via data models requires the use of a closed world assumption. To enable this the OWL syntax is still used but is interpreted in a closed world manner. For example, where OWL2 models domains of a property in order to infer the class of a certain entity, Discovery uses the same syntax for use in editorial policies. Where OWL2 may say that one of the domains of a causative agent is an allergy (i.e.an unknown class with a property of causative agent is likely to be an allergy), in the data modelling the editorial policy states that an allergy can only have properties that are allowed via the property domain. Thus the Snomed MRCM can be modelled in OWL2.
GRAPHQL
Graph QL , despite its name is not in itself a query language but a way of representing the graph like structure of a underlying model that has been built using OWL. GRAPH QL has a very simple class property representation, is ideal for REST APIs and results are JSON objects in line with the approach taken by the above Discovery syntax.
Nevertheless, GRAPHQL considers properties to be functions (high order logic) and therefore properties can accept parameters. For example, a patient's average systolic blood pressure reading could be considered a property with a single parameter being a list of the last 3 blood pressure readings.
Thus GRAPHQL capability is extended by enabling property parameters as types to support such things as filtering, sorting and limiting in the same way as any other query language by modelling types passed as parameters. Subqueries are then supported in the same way.
GRAPH QL has been chosen over SPARQL for reasons of simplicity and may now consider GRAPHQL to be a de-facto standard.
ABAC language
Main article : Discovery ABAC language
The standard XACML specifies a language that may be used to implement ABAC. XACML includes a set of grammatical concepts such as policy sets, policies, rules, combination rules, targets, obligations, effects and so on with many and variable sophisticated tokens and functions used to build the policy rules. XACML has its own XML syntax that can be used directly.
This language is somewhat disconnected with the other standards in terms of syntax and approach to vocab. Consequently Discovery uses a JSON profile of XACML as its ABAC language which itself models the attributes as OWL properties, and uses SPARQL as its rule representation.
Objectives and purposes
The purpose of the language is to help build a health information information model in a way that supports implementations of the model using different data base technologies and query languages.
The underlying philosophy behind the use language is described in the article : Information modelling language - philosophy
This article focuses on the description of the language itself.
As mentioned above each sublanguage is based on the grammar of a single recognised standards based language, the language having been selected as the ones that are the closest fit to the information requirements that the Discovery health information model is designed to support. A single grammar and optional single syntax enables the model to operate in an integrated manner, but at the same time enables the sublanguages to be represented in their native standard languages.
Like UMLS, the sublanguages use a common link in cross reference, a concept, which is identified with a unique identifier (Internationalised resource identifier - IRI) . a concept is usually named and defined semantically, and forms the means of traversing the model from different starting points to different end points, for different purposes.
Semantic Ontology
Main article Discovery semantic ontology language
The semantic ontology language is part of the Discovery information modelling language.
The grammar for the semantic ontology language used for the Discovery ontology is OWL EL, which is limited profile of OWL DL. The language used for data modelling and value set modelling is OWL2 DL as the more expressive constructs are required.
As such the ontology supports the OWL2 syntaxes such as the Functional syntax and Manchester syntax, but also supports the Discovery JSON based syntax, as part of the full information modelling language.
Together with the query language, OWL2 DL makes the language compatible also with Expression constraint language which is used as the standard for specifying Snomed-CT expression query.
Ontology purists will notice that modelling a data model in OWL2 is in fact a breach of the fundamental open world assumption view of the world taken in ontologies and instead applies the closed world assumption view instead. Consequently, a data model would normally be used independently of DL
The ontologies that are modelled are considered as modular ontologies. it is not expected that one "mega ontology" would be authored but that there would be maximum sharing of concept definitions (known as axioms) which results in a super ontology of modular ontologies.
Data modelling and semantic interoperability
Data models, and concept definitions and objects are modelled in OQL language using the Graph paradigm. As a result, all content can be viewed as semantic triples consisting of subject predicate and object.
Data Modelling takes account of ontology modularisation. A particular data model is a particular business oriented perspective on a set of concepts. As there are potentially thousands of different perspectives (e.g. a GP versus a geneticist) there are potentially unlimited number of data models. All the data models in Discovery share the same atomic concepts and same semantic definition across ontologies where possible, but where not, mapping relationships are used. The binding of a data model to its property values is based on a business specific model. For example a standard FHIR resource will map directly to the equivalent data model class, property and value set, whose meaning is defined in the semantic ontology, but the same data may be carried in a non FHIR resource without loss of interoperability.
A common approach to modelling and use of a standard approach to ontology, together with modularisation, means that any sending or receiving machine which uses concepts from the "super" ontology can adopt full semantic interoperability. If both machines use the same data model for the same business, the data may presented in the same relationship, but if the two machines use different data models for different businesses they may present the data in different ways but without any loss of meaning or query capability
Data mapping sublanguage
This part of the language is used to define mappings between the data model and an actual schema to enable query and filers to automatically cope with the ever extending ontology and data properties.
The language can be used to auto generate starter schemas for implementation i.e. schemas that will then be optimised for real world use.
the main use case for he mapping sublanguage is data transformation. This uses techniques such as Object relational mapping and therefore the transform instructions in the form of maps, follow this approach. There is no single standard for ORM maps but best practice of the kind supported by open source utilities such as Hibernate is followed: